Release Notes
Release notes information is provided in a scrolling format below. Each version heading is linked to a PDF of the corresponding releases notes. Click the link to download the PDF.
For information on contacting Tech Support, see Technical support contact information.
For copyright information, see Legal notices and copyright information.
New Features
The following features are available in BR.io for the 1.11 release.
Data Sharing Reviewer Role
BR.io Data Sharing now includes a Reviewer role. Users assigned to projects as a Reviewer can:
|
●
|
View CFX Opus and PTC Tempo files (protocols, runs, and templates) |
|
●
|
Download and export data from BR.io |
|
●
|
Download files from BR.io to your local device or computer for offline analysis and storage |
Project Admins can assign the Reviewer role to users, granting them view-only access to shared projects while preventing them from editing or adding data.
New Data Sharing Role Icons
Data Sharing project roles (Admin, Contributor, Reviewer) are represented by the following icons, depending on the role assigned to the user in a project:
|
●
|
Admin —  |
|
●
|
Contributor —  |
|
●
|
Reviewer —  |
These icons indicate a user's role in a project. They appear in the project selector in the upper-right corner of the BR.io page after project assignment and in the project destination dialog when a user moves files.
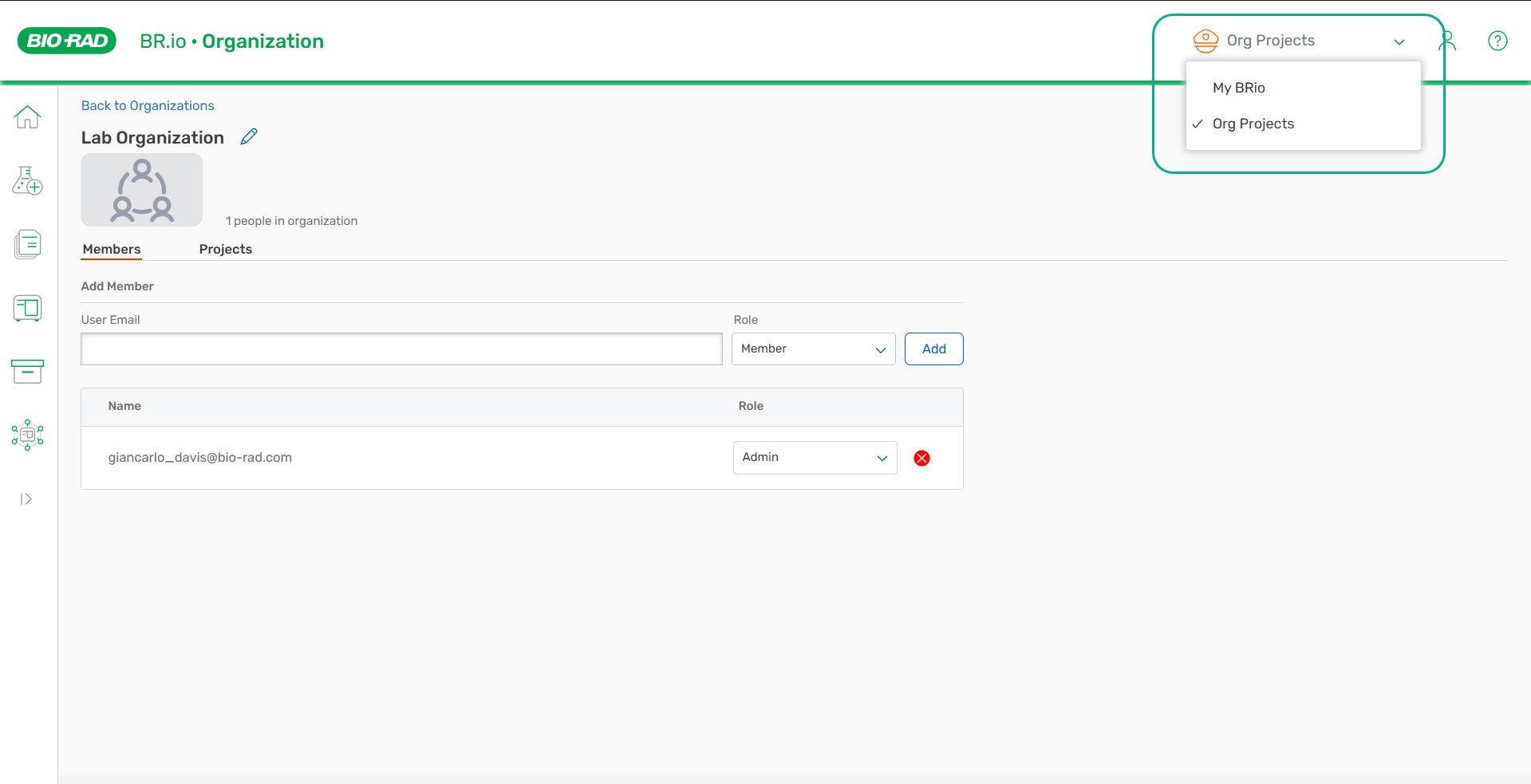
My BRio and Data Sharing Project Dropdown List Location
The My BRio folder and project folders are moved from the upper left to the upper right of any BR.io page to improve visibility and accessibility.
New Features
The following features are available in BR.io for the 1.10 release.
Fleet Management
BR.io now includes the Fleet Management feature, enabling users to reserve one or more PTC Tempo instruments to:
|
●
|
Assign protocols to instruments |
|
●
|
Open and close instrument lids |
|
●
|
Start and stop protocol runs as well as skip steps during a run on one or more instruments |
Note: Fleet Management is currently not available for PTC Tempo 48/48, CFX Opus, and ChemiDoc Go.
Important: If any changes you make in Fleet Management aren’t appearing (e.g., an instrument reservation, protocol assignment, or instrument control command), refresh the page to resolve any browser-related issues.
Known Issues
BR.io
|
●
|
If the PTC Tempo protocol that you want to assign to a reserved instrument is missing from the Manage Reservations protocol dropdown list, verify that the protocol appears in the PTC Protocols page. To access this page, click the Create icon in the left navigation bar, then click Manage PTC Protocols. |
|
●
|
BR.io might deny a reservation request even if the system shows the instrument status as "Available” because more than one user is submitting a request simultaneously. |
|
●
|
If you have created an organization project or are assigned an organization project, you will not be able to access those project protocols from the My Brio dropdown if you are on any of the Fleet Management pages. You must navigate out of the Fleet Management page to access those files. |
|
●
|
Reserved instruments that complete a run of an assigned protocol might not automatically clear the protocol assignment. If this occurs, remove the assigned protocol or assign a different one. |
|
●
|
The instrument Status column will always display “Connecting” if you click the Fleet Management icon after navigating out of Fleet Management or logging in to BR.io. |
|
●
|
The instrument Status column might intermittently display "Connecting" for all instruments connected to BR.io when you is on any Fleet Management page. However, this does not affect instrument connectivity to BR.io. |
|
●
|
The instrument Status column might continuously display "Connecting" when you navigate to the Fleet Management page and wait for the instrument status to load. If this occurs, either |
|
○
|
Click on any icon in the left navigation bar to navigate to another BR.io page, then return to Fleet Management. |
|
●
|
If your computer's time is not synchronized with official time, you cannot assign a protocol. Ensure that your computer's "Set Time and Date Automatically" setting is enabled, or restart the computer. |
|
●
|
You must refresh the BR.io web page after upgrading your PTC Tempo instrument to version 3.0 for it to appear in BR.io and to enable its functionality with Fleet Management. |
New Features
The following features are available in BR.io for the 1.9.1 release.
ChemiDoc Go Image Display and Details (Metadata) Enhancements
The ChemiDoc Go run page now displays the image captured from the ChemiDoc Go instrument. You can also select the image’s acquisition and image information as well as any notes by clicking the Acquisition Info, Image Info, or Notes buttons. For multichannel images, you can use arrows on either side of the image display to navigate between images and their corresponding information and notes.
New Features
The following features are available in BR.io for the 1.9 release.
Data Sharing
BR.io now includes a data sharing feature. BR.io data sharing makes the same data resources (such as protocols and protocol runs) available to multiple users within a designated group. Data sharing fosters collaboration among users, allowing authorized parties to leverage valuable research findings while safeguarding confidential information from unauthorized access.
Data sharing enables you to:
|
●
|
Create an organization, which includes a group of users (members) that can access the same data resources (CFX Opus protocols and runs and PTC Tempo completed runs) |
|
●
|
Add members to an organization and change their role as an organization admin or member |
|
●
|
Add projects to an organization. A project includes files that authorized members of an organization can access |
|
●
|
Assign members to projects |
|
●
|
Change project member roles to a project admin or a member |
Note: This feature is currently not available for ChemiDoc Go.
Known Issues
BR.io
|
●
|
If you open a file in BR.io before first selecting its organization, the file will be read-only. To edit and save the file, open the file from its organization on the organization list page. |
|
●
|
If two users from different organizations are working on separate projects and share a project file link, the first user might momentarily see the second user's project files before their own. This occurs when the first user closes the shared file tab after second user reviews the first user’s file. |
New Features
The following features are available in BR.io for the 1.8 release.
Integration with the ChemiDoc Go Imaging System
The ChemiDoc Go Imaging System facilitates image acquisition for selected gel and blot applications. Using Image Lab Touch from the instrument touch screen, you can acquire data from a single channel image or a multichannel image (1–3 channels) that can include fluorescence, chemiluminescence, and colorimetric applications.
After image data is acquired, you can
|
●
|
Export the image file to BR.io |
All images exported to BR.io appear on the Files page. An abbreviated list appears under Recent Files on the Home page.
|
●
|
Display the image details (metadata) in BR.io |
Click the image name to open the ChemiDoc Go Run page, which contains acquisition and image information.
|
●
|
Download the image file for viewing and analysis in Image Lab Software |
Image Lab Software (Version 6.1 or later) is Bio-Rad’s standalone software application for sophisticated image analysis. In Image Lab, you can display and analyze single channel and multi-channel gel and blot images. For more information, go to bio-rad.com.
Known Issues
BR.io
|
●
|
In the protocol view, if a protocol name with 255 characters contains no spaces, then the name does not fit in the text box. To avoid text overflow past the field boundaries, use spaces in long protocol names. |
|
●
|
You must perform a hard refresh (CTRL-F5 for Windows, CMD-SHIFT-R for a Mac) to display newly released online help content. |
|
●
|
BR.io accepts multiple ChemiDoc Go user accounts and allows them to link to the same BR.io user account; however, if one user unlinks the instrument from the BR.io account the instrument is removed from the BR.io Instruments page, but other users still see their accounts as linked on the instrument. To avoid confusion, Bio-Rad recommends that you link each instrument user account to a separate user account in BR.io. |
PTC Tempo
|
●
|
You might encounter the following issues when linking your PTC Tempo instrument to BR.io: |
|
○
|
If your browser is not connected to the network, submitting a valid instrument linking code prompts an invalid code error in BR.io. |
|
○
|
You must use lower-case characters for your BR.io username, or the login fails during the linking process. |
|
●
|
If your PTC Tempo instrument loses internet connectivity, an in-progress run remains in progress in BR.io, and the instrument fails to upload a run report. |
|
●
|
If you are using a Safari browser and the PTC Tempo run report file name is between 252 and 255 characters, you cannot download the run report as a PDF. You must rename the file with fewer characters or use Google Chrome to download the PDF. |
|
●
|
For content allowed in certain fields, note the following differences between BR.io and the PTC Tempo: |
Maximum time: In BR.io, you can set a maximum time for a protocol at 64800 seconds, but the PTC Tempo allows up to 64799 seconds only. If you enter 64800, the instrument automatically decreases the run time by 1 second.
Non-GOTO step fields: In BR.io, you can edit the Increment and Extend fields in temperature and gradient steps; however, the instrument does not allow editing of those fields and ignores the BR.io values during the run.
GOTO step counts: In BR.io, you can specify up to 9999 GOTO steps in a PTC Tempo protocol, but the instrument limits the user to 999 GOTO steps only. If you enter 1000 or more, the instrument automatically reduces the GOTO repetitions to 999.
Protocol name: In BR.io, you can name a PTC Tempo protocol using up to 255 characters, but the instrument allows up to 32 characters only.
Note: BR.io allows longer entries in alphanumeric character fields to support current and future connected instruments.
CFX Opus
|
●
|
BR.io allows you to create CFX Opus protocols containing melt and temperature steps under 4° C, although those temperatures are not supported by CFX Opus instruments. To avoid issues with the run, ensure the temperature is 4° C or above. |
|
●
|
You must close the run successfully uploaded to your BR.io account dialog box soon after the run is completed, or BR.io incorrectly displays the CFX Opus status as Offline. |
|
●
|
If you navigate from the CFX run workflow while uploading a file, BR.io does not warn you about unsaved changes to your CFX run. |
|
●
|
For CFX Opus templates, note the following: |
|
○
|
You must create CFX Opus run templates from an existing completed CFX run file. |
|
○
|
You cannot open, view, or edit CFX Opus run templates independently. |
|
○
|
After saving the run file created from a template, you cannot directly edit the sample list. |
|
○
|
If the sample list contains fewer samples than the plate layout accepts, you must open the run file after it has been saved, and manually clear the unused wells from the plate. |
|
●
|
BR.io does not currently support the following: |
|
○
|
Analysis of .pcrd or .zpcr files containing legacy or user-calibrated fluorophores; you can upload the files in BR.io, but errors are likely if you work with the data in the Analysis module. |
|
○
|
Application-based analysis, such as standard curve/absolute quantification, gene expression/relative quantification, and allelic discrimination |
New Features
The following features are available in BR.io for the 1.7.1 release.
Connections to PTC Tempo 384-Well and 48/48 Dual Block Thermal Cyclers
In addition to PTC Tempo thermal cyclers with 96-well and 96-deepwell blocks, you can connect your 384-well block and 48/48 dual block PTC Tempo thermal cyclers to BR.io.
PTC Tempo Instrument Status Icons
You can view the following PTC Tempo instrument statuses in the Home and Instrument pages in BR.io:

|
Idle |

|
Offline |

|
Connecting |

|
Running |

|
Paused |

|
Infinite Hold |

|
Error |
Important: You might need to update your PTC Tempo software and firmware to view the different instrument statuses that appear in the Status column in BR.io. Click the link below and scroll through the available downloads in the Download section to locate the PTC Tempo software and firmware.
https://www.bio-rad.com/pcrupdates
Log into the website and follow the prompts to download the installation files.
PTC Tempo Infinite Hold Option
An Infinite Hold status option is available for temperature and gradient steps in PTC Tempo protocols. Infinite Hold automatically holds a defined temperature at a particular step in the protocol until the hold is manually released. You can add an Infinite Hold step from BR.io or from your PTC Tempo thermal cycler.
Improvements
New Locations for Upload Icon
The Upload icon has been removed from the application banner and relocated to the secondary banner in the Files and Manage Protocols pages.
Known Issues
BR.io
|
●
|
In the protocol view, if a protocol name with 255 characters contains no spaces, then the name does not fit in the text box. To avoid text overflow past the field boundaries, use spaces in long protocol names. |
PTC Tempo
|
●
|
You might encounter the following issues when linking your PTC Tempo instrument to BR.io: |
|
○
|
If your browser is not connected to the network, submitting a valid instrument linking code prompts an invalid code error in BR.io. |
|
○
|
You must use lower-case characters for your BR.io username, or the login fails during the linking process. |
|
●
|
If your PTC Tempo instrument loses internet connectivity, an in-progress run remains in progress in BR.io, and the instrument fails to upload a run report. |
|
●
|
If you are using a Safari browser and the PTC Tempo run report file name is between 252 and 255 characters, you cannot download the run report as a PDF. You must rename the file with fewer characters or use Google Chrome to download the PDF. |
|
●
|
For content allowed in certain fields, note the following differences between BR.io and the PTC Tempo: |
Maximum time: In BR.io, you can set a maximum time for a protocol at 64800 seconds, but the PTC Tempo allows up to 64799 seconds only. If you enter 64800, the instrument automatically decreases the run time by 1 second.
Non-GOTO step fields: In BR.io, you can edit the Increment and Extend fields in temperature and gradient steps; however, the instrument does not allow editing of those fields and ignores the BR.io values during the run.
GOTO step counts: In BR.io, you can specify up to 9999 GOTO steps in a PTC Tempo protocol, but the instrument limits the user to 999 GOTO steps only. If you enter 1000 or more, the instrument automatically reduces the GOTO repetitions to 999.
Protocol name: In BR.io, you can name a PTC Tempo protocol using up to 255 characters, but the instrument allows up to 32 characters only.
Note: BR.io allows longer entries in alphanumeric character fields to support current and future connected instruments.
CFX Opus
|
●
|
BR.io allows you to create CFX Opus protocols containing melt and temperature steps under 4° C, although those temperatures are not supported by CFX Opus instruments. To avoid issues with the run, ensure the temperature is 4° C or above. |
|
●
|
You must close the run successfully uploaded to your BR.io account dialog box soon after the run is completed, or BR.io incorrectly displays the CFX Opus status as Offline. |
|
●
|
If you navigate from the CFX run workflow while uploading a file, BR.io does not warn you about unsaved changes to your CFX run. |
|
●
|
For CFX Opus templates, note the following: |
|
○
|
You must create CFX Opus run templates from an existing completed CFX run file. |
|
○
|
You cannot open, view, or edit CFX Opus run templates independently. |
|
○
|
After saving the run file created from a template, you cannot directly edit the sample list. |
|
○
|
If the sample list contains fewer samples than the plate layout accepts, you must open the run file after it has been saved, and manually clear the unused wells from the plate. |
|
●
|
BR.io does not currently support the following: |
|
○
|
Analysis of .pcrd or .zpcr files containing legacy or user-calibrated fluorophores; you can upload the files in BR.io, but errors are likely if you work with the data in the Analysis module. |
|
○
|
Application-based analysis, such as standard curve/absolute quantification, gene expression/relative quantification, and allelic discrimination |
Summary of PTC Tempo Features
In version 1.7, BR.io is integrated with the PTC Tempo conventional PCR instrument, allowing you to do the following:
|
●
|
Follow the process from the instrument to BR.io to link your PTC Tempo 96-well and 96-deepwell instruments to your BR.io user account |
|
●
|
Create protocols in BR.io for the 96-well or 96-deepwell systems |
|
●
|
View and run your BR.io protocols on linked PTC Tempo instruments |
|
●
|
View run data that is automatically uploaded to BR.io upon completion of the PTC Tempo run |
The following sections describe in more detail the capabilities of PTC Tempo instruments linked to BR.io.
Linking Your Instruments
|
●
|
Link PTC Tempo instruments using OAuth 2.0's code flow for authorized access |
Managing Linked Instruments in BR.io
|
●
|
View your linked instruments on the Instruments page, and recently used instruments as icons on the BR.io Home page |
|
●
|
On the BR.io Instruments page, view instrument details for your linked PTC Tempo (for example, serial number, instrument name and model, and software and firmware versions) |
Creating and Managing PTC Tempo Protocols in BR.io
|
●
|
BR.io provides a dedicated repository for your protocols, separated by instrument |
|
●
|
You can create, edit, and archive protocols in BR.io |
|
●
|
Protocols in BR.io appear in both graphical step format and a protocol step list |
|
●
|
You can add or remove temperature, gradient, and GOTO steps in your PTC Tempo protocols |
|
●
|
Protocol parameters are validated, and error messages appear where applicable |
Viewing Email Notifications, Completed Run Details, and Run Report
|
●
|
You can receive and view completed run information, as follows: |
|
●
|
Receive an email notification when your run on the PTC Tempo is completed |
|
●
|
In BR.io, access a summary of your completed run in BR.io and open a run report containing temperatures and durations for the protocol steps |
|
●
|
From Brio, download the run report in PDF format |
Interoperability with Legacy Instruments
|
●
|
You can upload and export protocol files in .prcl format, which can be used on legacy instruments |
Fixed Issues
The following issues with the CFX Opus have been resolved:
|
●
|
The correct CFX Opus instrument is now identified in the subject line and message body in email notifications for completed CFX Opus runs. |
|
●
|
If there is a protocol mismatch between CFX Opus and BR.io, an internal error appears and you cannot proceed with the run until the error is corrected. To proceed with the run, the user must do the following: |
1. Exit out of the run on the instrument.
2. Refresh the file browser on the instrument to get the latest version of the pending run.
3. Reload the run before starting it.
Known Issues
BR.io
|
●
|
In the protocol view, if a protocol name with 255 characters contains no spaces, then the name does not fit in the text box. To avoid text overflow past the field boundaries, use spaces in long protocol names. |
PTC Tempo
|
●
|
You might encounter the following issues when linking your PTC Tempo instrument to BR.io: |
|
○
|
If your browser is not connected to the network, submitting a valid instrument linking code prompts an invalid code error in BR.io. |
|
○
|
You must use lower-case characters for your BR.io username, or the login fails during the linking process. |
|
●
|
If your PTC Tempo instrument loses internet connectivity, an in-progress run remains in progress in BR.io, and the instrument fails to upload a run report. |
|
●
|
If you are using a Safari browser and the PTC Tempo run report file name is between 252 and 255 characters, you cannot download the run report as a PDF. You must rename the file with fewer characters or use Google Chrome to download the PDF. |
|
●
|
For content allowed in certain fields, note the following differences between BR.io and the PTC Tempo: |
Maximum time: In BR.io, you can set a maximum time for a protocol at 64800 seconds, but the PTC Tempo allows up to 64799 seconds only. If you enter 64800, the instrument automatically decreases the run time by 1 second.
Non-GOTO step fields: In BR.io, you can edit the Increment and Extend fields in temperature and gradient steps; however, the instrument does not allow editing of those fields and ignores the BR.io values during the run.
GOTO step counts: In BR.io, you can specify up to 9999 GOTO steps in a PTC Tempo protocol, but the instrument limits the user to 999 GOTO steps only. If you enter 1000 or more, the instrument automatically reduces the GOTO repetitions to 999.
Protocol name: In BR.io, you can name a PTC Tempo protocol using up to 255 characters, but the instrument allows up to 32 characters only.
Note: BR.io allows longer entries in alphanumeric character fields to support current and future connected instruments.
CFX Opus
|
●
|
BR.io allows you to create CFX Opus protocols containing melt and temperature steps under 4° C, although those temperatures are not supported by CFX Opus instruments. To avoid issues with the run, ensure the temperature is 4° C or above. |
|
●
|
You must close the run successfully uploaded to your BR.io account dialog box soon after the run is completed, or BR.io incorrectly displays the CFX Opus status as Offline. |
|
●
|
If you navigate from the CFX run workflow while uploading a file, BR.io does not warn you about unsaved changes to your CFX run. |
|
●
|
For CFX Opus templates, note the following: |
|
○
|
You must create CFX Opus run templates from an existing completed CFX run file. |
|
○
|
You cannot open, view, or edit CFX Opus run templates independently. |
|
○
|
After saving the run file created from a template, you cannot directly edit the sample list. |
|
○
|
If the sample list contains fewer samples than the plate layout accepts, you must open the run file after it has been saved, and manually clear the unused wells from the plate. |
|
●
|
BR.io does not currently support the following: |
|
○
|
Analysis of .pcrd or .zpcr files containing legacy or user-calibrated fluorophores; you can upload the files in BR.io, but errors are likely if you work with the data in the Analysis module. |
|
○
|
Application-based analysis, such as standard curve/absolute quantification, gene expression/relative quantification, and allelic discrimination |
Summary of New Features
New User Account Verification
When a user signs up for a BR.io account, the application prompts the user to confirm the email address before the account is activated.
|
●
|
After clicking Create Account, BR.io displays a welcome message that advises the user to look for the account verification email in the email Inbox. Users should also check their Spam or Junk folder if the email does not arrive in their Inbox within a few minutes. |
|
●
|
When the user opens the email and clicks the verification hyperlink, the account is verified and the BR.io login page opens, with a “Verified” confirmation above the login fields. If the link in the email has expired, the user can request a new verification link. Only the most recently issued link is valid. |
Known Issues
|
●
|
You must close the Run successfully uploaded to your BR.io account dialog box soon after the run is completed, or BR.io incorrectly displays the CFX Opus status as Offline. |
|
●
|
If you navigate from the CFX run workflow while uploading a file, BR.io does not warn you about unsaved changes to your CFX run. |
|
●
|
You must create CFX Opus run templates from an existing completed CFX run file. |
|
●
|
You cannot open, view, or edit CFX Opus run templates independently. |
|
●
|
After saving the run file created from a template, you cannot directly edit the sample list. |
|
●
|
If the sample list contains fewer samples than the plate layout accepts, you must open the run file after it has been saved, and manually clear the unused wells from the plate. |
|
●
|
If you change the Scan Mode for a pending CFX run in BR.io after loading it on the instrument the run will be performed using the original scan mode. |
Workaround: Exit out of the run on the instrument, refresh the file browser on the instrument, then reload the run before starting it.
|
●
|
Email notifications for runs completed on the Opus Deepwell instrument incorrectly state in the email subject line and body that the run was completed on the CFX Opus 384 instrument instead of the deepwell instrument. The email has no impact on instrument functionality or the run file. |
|
●
|
BR.io does not currently support |
|
○
|
Analysis of .pcrd or .zpcr files that contain legacy or user-calibrated fluorophores |
Note: You can upload the files, but working with them in the Analysis module can produce errors.
|
○
|
Application-based analysis, such as standard curve/absolute quantification, gene expression/relative quantification, and allelic discrimination |
Summary of New Features
Support for CFX Opus Deepwell System
The BR.io Cloud Platform now supports the CFX Opus Deepwell System, as follows:
|
●
|
To create a run for a CFX Opus Deepwell System, set the Plate Size to 96 Wells. You can use all CFX protocols created in BR.io with a CFX Opus Deepwell System. |
|
●
|
When adding a gradient step to a CFX protocol, select the 96D button to display the gradient for a CFX Opus Deepwell System. |
Fixed Issues
|
●
|
A maximum number of 500 BR.io users can be linked to a CFX Opus instrument (previously 20). |
|
●
|
Exported .pcrd files that used a protocol containing a gradient step and were opened in CFX Maestro Software displayed the incorrect gradient. |
Known Issues
|
●
|
You must close the Run successfully uploaded to your BR.io account dialog box soon after the run is completed, or BR.io incorrectly displays the CFX Opus status as Offline. |
|
●
|
If you navigate from the CFX run workflow while uploading a file, BR.io does not warn you about unsaved changes to your CFX run. |
|
●
|
You must create CFX Opus run templates from an existing completed CFX run file. |
|
●
|
You cannot open, view, or edit CFX Opus run templates independently. |
|
●
|
After saving the run file created from a template, you cannot directly edit the sample list. |
|
●
|
If the sample list contains fewer samples than the plate layout accepts, you must open the run file after it has been saved, and manually clear the unused wells from the plate. |
|
●
|
If you change the Scan Mode for a pending CFX run in BR.io after loading it on the instrument the run will be performed using the original scan mode. |
Workaround: Exit out of the run on the instrument, refresh the file browser on the instrument, then reload the run before starting it.
|
●
|
BR.io does not currently support |
|
○
|
Analysis of .pcrd or .zpcr files that contain legacy or user-calibrated fluorophores |
Note: You can upload the files, but working with them in the Analysis module can produce errors.
|
○
|
Application-based analysis, such as standard curve/absolute quantification, gene expression/relative quantification, and allelic discrimination |
Summary of New Features
Email Notifications for Completed CFX Opus Runs
|
●
|
You can choose to receive notifications by email when your run is completed on a CFX Opus system. |
Fixed Issues
|
●
|
When you change a protocol for a pending CFX run in BR.io after it is loaded on the CFX Opus system, you cannot start the run. |
|
○
|
Refresh the file browser and load the run again to override the error state and start the run. |
Known Issues
|
●
|
You must close the “Run successfully uploaded to your BR.io account” dialog box soon after the run is completed, or BR.io incorrectly displays the CFX Opus status as Offline. |
|
●
|
If you navigate from the CFX run workflow while uploading a file, BR.io does not warn you about unsaved changes to your CFX run. |
|
●
|
CFX Opus run templates must be created from an existing completed CFX run file. |
|
●
|
CFX Opus run templates cannot be opened, viewed, or edited independently. |
|
●
|
After saving the run file created from a template, you cannot directly edit the sample list. |
|
●
|
If the sample list contains fewer samples than the plate layout accepts, you must open the run file after it has been saved, and manually clear the unused wells from the plate. |
|
●
|
If you change the Scan Mode for a pending CFX run in BR.io after loading it on the instrument, the run uses the original scan mode. |
Workaround: Exit out of the run on the instrument, refresh the file browser on the instrument, then reload the run before starting it.
|
●
|
BR.io does not currently support |
|
○
|
Analysis of .pcrd or .zpcr files that contain legacy or user-calibrated fluorophores |
Note: You can upload the files, but working with them in the Analysis module can produce errors.
|
○
|
Application-based analysis, such as standard curve/absolute quantification, gene expression/relative quantification, and allelic discrimination |
Summary of New Features
File Search
|
●
|
You can search for particular protocols, run files, and templates using the Search field on the Files page. |
Fluorescence Drift Correction
|
●
|
From the options in the Analysis Setup screen, you can apply fluorescence drift correction to your completed run data. |
Fixed Issues
|
●
|
CFX protocols that are restored from the archive may not appear in the Select Protocol dropdown menu when creating a CFX run until the page is refreshed. |
Known Issues
|
●
|
You must close the “Run successfully uploaded to your BR.io account” dialog box soon after the run is completed, or BR.io incorrectly displays the CFX Opus status as Offline. |
|
●
|
If you navigate from the CFX run workflow while uploading a file, BR.io does not warn you about unsaved changes to your CFX run. |
|
●
|
CFX Opus run templates must be created from an existing completed CFX run file. |
|
●
|
CFX Opus run templates cannot be opened, viewed, or edited independently. |
|
●
|
After saving the run file created from a template, you cannot directly edit the sample list. |
|
●
|
If the sample list contains fewer samples than the plate layout accepts, you must open the run file after it has been saved, and manually clear the unused wells from the plate. |
|
●
|
BR.io does not currently support |
|
○
|
Analysis of .pcrd or .zpcr files that contain legacy or user-calibrated fluorophores |
Note: Note: You can upload the files, but working with them in the Analysis module can produce errors.
|
○
|
Application-based analysis, such as standard curve/absolute quantification, gene expression/relative quantification, and allelic discrimination |
Summary of New Features
FRET Scan Mode
|
●
|
You can create and execute CFX Opus runs using the FRET scan mode, and you can open and analyze completed CFX runs that use the FRET scan mode. |
Note: This applies to CFX runs set up in BR.io and CFX runs manually uploaded as a .pcrd or .zpcr file.
Baseline Cycle Range
|
●
|
You can now adjust the cycle range that is used to determine the baseline for baseline subtraction. |
Run Templates
|
●
|
You can now create run templates that enable you to quickly set up CFX Opus runs using predefined protocol, plate layout, and analysis settings. Templates also provide a streamlined workflow for entering the sample list for the run. |
Early Access Program Sign-Up
|
●
|
You can now express interest in joining the BR.io Early Access Program. From the Explore BR.io panel on the Home page, click Join the Early Access Program. |
Known Issues
|
●
|
You must close the “Run successfully uploaded to your BR.io account” dialog box soon after the run is completed, or BR.io incorrectly displays the CFX Opus status as Offline. |
|
●
|
If you navigate from the CFX run workflow while uploading a file, BR.io does not warn you about unsaved changes to your CFX run. |
|
●
|
CFX protocols that are restored from the archive may not appear in the Select Protocol dropdown menu when creating a CFX run until the page is refreshed. |
|
●
|
CFX Opus run templates must be created from an existing completed CFX run file. |
|
●
|
CFX Opus run templates cannot be opened, viewed, or edited independently. |
|
●
|
After saving the run file created from a template, you cannot directly edit the sample list. |
|
●
|
If the sample list contains fewer samples than the plate layout accepts, you must open the run file after it has been saved, and manually clear the unused wells from the plate. |
|
●
|
BR.io does not currently support |
|
○
|
Analysis of .pcrd or .zpcr files that contain legacy or user-calibrated fluorophores |
Note: You can upload the files, but working with them in the Analysis module can produce errors.
|
○
|
Fluorescence drift correction analysis settings |
|
○
|
Application-based analysis, such as standard curve/absolute quantification, gene expression/relative quantification, and allelic discrimination |
Summary of New Features
Gradient Calculator for CFX Protocol Creation
|
●
|
When you add or edit a gradient step in a CFX protocol, you can view a gradient calculator that displays the temperature for each row of the plate. |
New Plate Functionality for CFX Run Creation
|
●
|
You can copy a plate layout from an existing CFX run into your new CFX run. |
|
●
|
Row and column identifiers remain visible when you scroll through the plate. |
|
●
|
New target colors provide better contrast while you are editing the plate. |
Additional Functionality in CFX Run Analysis
|
●
|
New functionality for CFX melt curve analysis: |
|
○
|
You can view the melt curve chart. |
|
○
|
You can set a manual threshold in the melt peak chart. |
|
●
|
Row and column identifiers remain visible when you scroll through the plate layout. |
Additional CFX Run Details
|
●
|
On the Run page you can |
|
○
|
View the date and time of a completed CFX run |
|
○
|
View details about the instrument used |
|
○
|
Open a run log for the completed CFX run |
CFX Maestro Interoperability
|
●
|
You can export completed CFX runs in a .pcrd file format for analysis in the CFX Maestro Software. |
User Management
|
●
|
For validation purposes when creating a new BR.io account, user must reenter their email address and password. |
Archiving and Restoring Files
|
●
|
BR.io features an Archive page, accessible from the main navigation panel, when you can |
|
○
|
Archive files and protocols instead of permanently deleting them |
|
○
|
View the list of archived files |
Note: You must restore a file before you can open it in BR.io.
Improved Error Messages
|
●
|
Error messages throughout BR.io have been improved for better clarity. |
BR.io on Mobile Devices
|
●
|
The BR.io landing page is updated for better viewing on mobile devices. |
Contacting Technical Support
|
●
|
When you request assistance using the BR.io Contact Support feature, you can |
|
○
|
Select the region where your facility is located |
Fixed Issues
|
●
|
Users who mistype their email address during signup no longer experience issues with signing in or resetting their password. |
|
●
|
After you exit the Target Name field, BR.io does not assign a color to a new target on the Plate Setup for a CFX run. See information on new target colors in plate setup. |
|
●
|
Plate headers (row and column) remain in view on Plate Setup while you are scrolling. |
|
●
|
BR.io now supports the following: |
|
○
|
Viewing or analyzing in completed CFX runs with melt curve data, but without an amplification step. |
|
○
|
Viewing the gradient (temperature per row) when adding a Gradient step to a CFX protocol. |
|
○
|
Viewing details and metadata for a completed CFX run, including the date and time of the run, and the name, model, and serial number of the instrument. |
|
○
|
Downloading CFX runs created or modified in BR.io as CFX Maestro-compatible .pcrd files. |
|
●
|
Melt curve no longer fails to load. |
Known Issues
|
●
|
You must close the “Run successfully uploaded to your BR.io account” dialog box soon after the run is completed, or BR.io incorrectly displays the CFX Opus status as Offline. |
|
●
|
If you navigate from the CFX run workflow while uploading a file, BR.io does not warn you about unsaved changes to your CFX run. |
|
●
|
BR.io does not currently support |
|
○
|
Analysis of .pcrd or .zpcr files that contain legacy or user-calibrated fluorophores |
Note: You can upload the files, but working with them in the Analysis module can produce errors.
|
○
|
Baseline adjustment and fluorescence drift correction analysis settings |
|
○
|
Application-based analysis, such as standard curve/absolute quantification, gene expression/relative quantification, and allelic discrimination |
Summary of Main Features
Individual User Accounts
|
●
|
You can create an individual BR.io user account, remain logged in for up to 30 days, change a password, and reset a forgotten password. |
Online Assistance and Accessibility
|
●
|
BR.io features a Help Center that provides searchable software documentation and context-sensitive Help that is coordinated with primary BR.io pages. |
|
●
|
The BR.io interface is designed for ease-of-use by all users. |
|
●
|
Users can contact Bio-Rad Customer Support directly from BR.io. |
Storing and Managing Data
|
●
|
BR.io displays the full list of a user’s files on the Files page, and the most recently used files on the Home page. |
|
●
|
Users can upload .zpcr and .pcrd file types to the Files page, and rename and delete files. |
Integration with CFX Opus Instruments
|
●
|
Using the BR.io Cloud Platform you can |
|
○
|
Connect CFX Opus instruments to your BR.io account |
|
○
|
Create a run in BR.io, and then execute the run on the CFX Opus instrument |
|
○
|
Create a run on the CFX Opus instrument using a locally stored protocol, and save it to your storage area in BR.io |
|
●
|
For runs created or saved in BR.io, CFX Opus run data is automatically uploaded to BR.io from connected instruments. |
Managing and Monitoring CFX Opus Instruments
|
○
|
A list of all CFX Opus instruments connected to your BR.io account |
|
○
|
The model, name, and serial number of a connected CFX Opus instrument |
|
○
|
The current status, time remaining, and current step/cycle for an experiment running on a connected CFX Opus instrument |
Creating and Managing CFX Protocols
|
●
|
BR.io provides a dedicated repository for your protocols. |
|
●
|
You can create, edit, and delete protocols in BR.io. |
|
●
|
CFX protocols in BR.io appear in both a graphical step format and a list of protocol steps. |
|
●
|
In BR.io, you can create temperature, gradient, melt curve, and goto steps in your protocols. |
|
●
|
BR.io validates your protocol parameters and displays an error message if applicable. |
Creating and Editing a CFX Opus Run in BR.io
|
○
|
Streamlined workflow to set up an experiment to run on the CFX Opus 96 or CFX Opus 384 |
|
○
|
Plate editor that is optimized for usability, where you can see well content, select and edit multiple wells at once, and see plate validation and error messages, if applicable |
Note: BR.io only supports the following scan modes:
SYBR/FAM only
All channels
|
●
|
You can edit the plate and run details while the run is in progress, or after it has completed on the instrument. |
|
●
|
You can set up a CFX Opus run using a default (quick) plate. Default plates consist of pre-defined fluorophores and unknowns in every well, so you can skip or defer plate setup. |
BR.io Analysis Features
|
●
|
BR.io uses the same algorithms as CFX Maestro. |
|
●
|
BR.io enables the user to analyze real-time PCR (amplification) data, including the ability to |
|
○
|
Toggle baseline subtraction on or off |
|
○
|
Set automatic or user defined single thresholds |
|
○
|
View and download the amplification chart and apply linear or logarithmic scaling of the RFU data |
|
○
|
View amplification data in a sortable table format |
|
○
|
Export amplification data table and RFU data per cycle in .csv format |
|
○
|
Analyze data by fluorophore or target |
|
○
|
Show or hide data from individual wells in the plate |
|
○
|
Filter out targets or fluorophores from analysis |
|
●
|
Analyze melt curve data from CFX runs (Opus and legacy), including |
|
○
|
A melt peak (negative derivative) chart that you can download and print |
|
○
|
A sortable, exportable data table, which displays melting temperature, peak height, begin temperature, and end temperature |
Interoperability with CFX Maestro or CFX Manager
|
○
|
Upload and import .zpcr files and .pcrd files created in CFX Manager v1.0 and later, or CFX Maestro v1.0 and later |
|
○
|
Upload and import .pcrd files from CFX Maestro for Mac |
|
○
|
Download .zpcr file from a CFX Opus run |
Note: The plate layout and run details are omitted.
|
○
|
Download the original .zpcr or .pcrd file from a manually uploaded (imported) CFX run |
Known Issues
|
●
|
Users who mistype their email address during signup can experience issues with signing in or resetting their password. |
|
●
|
You must close the “Run successfully uploaded to your BR.io account” dialog box soon after the run is completed, or BR.io incorrectly displays the CFX Opus status as Offline. |
|
●
|
If you navigate from the CFX run workflow while uploading a file, BR.io does not warn you about unsaved changes to your CFX run. |
|
●
|
After you exit the Target Name field, BR.io assigns a color to a new target on the Plate Setup for a CFX run. |
|
●
|
Plate headers (row and column) do not remain in view on Plate Setup while scrolling. |
|
●
|
BR.io does not currently support |
|
○
|
Analysis of .pcrd or .zpcr files that contain legacy or user-calibrated fluorophores |
Note: You can upload the files, but working with them in the Analysis module can produce errors.
|
○
|
Viewing or analyzing completed CFX runs with melt curve data, but no amplification step |
|
○
|
Viewing the gradient (temperature per row) when adding a Gradient step to a CFX protocol |
|
○
|
Viewing details and metadata for a completed CFX run, including the date and time of the run, and the name, model and serial number of the instrument |
|
○
|
Downloading CFX runs created or modified in BR.io as CFX Maestro-compatible PCRD files |
|
○
|
Baseline adjustment and fluorescence drift correction analysis settings |
|
○
|
Application-based analysis, such as standard curve/absolute quantification, gene expression/relative quantification, and allelic discrimination |

